Overexpression and Prognostic Significance of PTPN2 May be a Novel Immunotherapy Target in Renal Clear Cell Carcinoma
Peipei Liu
Published Date: 2021-08-24DOI10.36648/2472-5056.21.s3.008
Peipei Liu*
Department of Histology and Embryology, Shenyang Medical College, Shenyang, China
- *Corresponding Author:
- Peipei Liu
Department of Histology and Embryology,
Shenyang Medical College,
Shenyang,
China,
E-mail: liupeipei@symc.edu.cn
Received Date: July 29, 2021l; Accepted Date: August 12, 2021;Published Date: August 19, 2021
Citation: Liu P (2021) Overexpression and Prognostic Significance of PTPN2 May be a Novel Immunotherapy Target in Renal Clear Cell Carcinoma. J Clin Exp Nephrol Vol. 6 No.S3: 008.
Abstract
Immunotherapy has significantly advanced in Clear Cell Renal Cell Carcinoma (ccRCC). We aimed to find a new immune-related prognostic biomarker and immunotherapeutic target for ccRCC. We analyzed the expression, survival, and related immune gene marker sets data of PTPN2 in patients with ccRCC from TCGA. PTPN2 expression was increased in ccRCC compared to normal tissue. PTPN2 was closely related to T stage (P=0.008), TNM stage (P=0.017) and Grade (P=0.002). Overexpression of PTPN2 predicted a poor survival in ccRCC (P< 0.001). PTPN2 was also related to six types of tumor immune-infiltrating cells, including B cells, CD8+T cells, CD4+T cells, Macrophage, Neutrophils, Dendritic cells. PTPN2 was related CTLA-4 (P=5.645404E-26, r=0.4339333) and PDCD1 (P=5.645404E-26, r=0.4339333). Furthermore, the survival rate in patients with high PTPN2 and CTLA4 was significantly lower than that other three strata (P<0.0001). GSEA and GO biological analysis was conducted, which was indicated PTPN2 was involved in many immune and inflammatory pathways, including IL-4, IL-12, CD3 T-cell, CD4 T-cell, intestinal inflammation, systemic inflammatory regulation related factors, and so on. Our results implied that PTPN2 was considered as the potential immune therapeutic target and prognostic biomarker in ccRCC.
Keywords
ARenal cell; Carcinoma; Immunotherapy; Prognosis; Protein tyrosine phosphatase; Non-receptor type 2
Introduction
As the most common pathological subtype of Renal Cell Carcinoma (RCC), Clear Cell Renal Cell Carcinoma (ccRCC) is associated with high morbidity and poor prognosis. To date, surgery is also the primary treatment for most ccRCC; radiotherapy and chemotherapy are largely ineffective in the treatment of ccRCC. To improve the survival of ccRCC, many Immune Checkpoint Inhibitors (ICIs) have been approved by the FDA, such as nivolumab and ipilimumab. However, only a few patients with advanced renal cancer have responded to immunotherapy. So the new immunotherapy target may improve the naïve vision of single target-based immunotherapy [1-6].
PTPN2 was discovered in T-cells, which is also known as T-Cell Protein Tyrosine Phosphatases (TCPTP). PTPN2 negatively regulated the pro-inflammatory pathways, such as INF-γ induced Janus kinase (JAK)-signaling and STAT signaling et al. The importance of PTPN2 in regulating tumorigenicity pathways were also highlighted [7]. The study showed that PTPN2 was negatively associated with activation of AKT in breast cancer. Grohmedann et al. also demonstrated that depletion of PTPN2 in hepatocytes promoted Hepatocellular Carcinoma (HCC) in mice. PTPN2 may play a tumor-suppressive role in tumors based on the above studies. A fascinating new study published in Nature has reported that PTPN2 deletion markedly increased the response of tumors to immunotherapy by enhancing interferon-γ-mediated effects on antigen presentation and growth suppression. Besides, Wiede et al. reported that PTPN2 could regulate the production of exhausted CD8 positive T cell subsets and control tumor immunity [8]. Due to the role of PTPN2 in tumor immunity, it may become a new target of immunotherapy [7-12].
Therefore, in this study, we firstly investigated the expression profiling and prognostic value of PTPN2 in various solid tumors based on TCGA datasets. Then we focused on Kidney Renal Clear Cell Carcinoma (KIRC) and tried to pinpoint the meaningful finding for future cancer immunotherapy. In this study, ccRCC was used instead of KIRC.
Materials and Methods
Expression profiling of PTPN2 in human cancers
The Tumor Immunological Estimation Resource (TIMER) platform was used to explore the expression profiling of PTPN2 in pan-cancer and the correlation between infiltration levels of six immune cell types (B cells, CD4+T cells, CD8+T cells, neutrophils, macrophages, and dendritic cells) in KIRC [23]. PTPN2 transcriptome data from 31 types of tumors in the TCGA dataset were obtained from the University of California, Santa Cruz Cancer. Gene expression level was presented as log2 RSEM (RNA-Seq by Expectation-Maximization). To determine the prognostic significance of PTPN2 in pan-cancer, the significance of Cox proportional hazards models were performed using the survival and survminer packages. The forest plot was performed with the forest plot package in the R language.
Correlations between clinic pathologic data and PTPN2 expression in KIRC
The clinic pathological information of Kidney Renal Clear Cell Carcinoma (KIRC), including gender, age, TNM stage, T stage, N stage, M stage. The correlations between clinicopathologic data and PTPN2 expression were analyzed by SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). Patients with KIRC were divided into high and low groups according to the median value of PTPN2 expression. The PTPN2 expression of different clinicopathologic groups was compared by Chiq test or student’s t-test. Overall survival was estimated by the Kaplan–Meier method and compared by log-rank tests using survival and survminer package in R language. Univariate and multivariate Cox regression analyses were used to compare the effect of PTPN2 on prognosis, along with the included clinical variables. A value of P<0:05 were considered statistically significant. Based on Cox multivariate regression analysis for OS, a nomogram was formulated by the package of rms package in R language.
Functional annotation of co-expression gene network of PTPN2
To explore the unique roles of PTPN2, PTPN2 co-expression genes were identified in the TCGA KIRC database with R language by setting the Pearson coefficient >0.4. Gene ontology (GO)/KEGG terms, canonical pathways, hall mark gene sets enrichment among the co-expression genes of PTPN2 was performed using Metascape [24]. The immunologically relevant list of genes curated with functions and Gene Ontology terms was downloading from the immunology database and analysis portal (ImmPort) system [25]. ImmPort integrates across publicly available datasets related to immunology data and facilitates transparency and reproducibility in immunology research.
Functional gene set enrichment analysis (GSEA) of PTPN2
GSEA was carried out using the java-based graphical user interface GSEA v4.1.0. For our research, PTPN2 expression levels in KIRC were dichotomized into two groups to annotate phenotype. The: C7 immunologic signature gene sets were downloaded from the Broad Institute Molecular Signature Database (MSigDB v7.1). To characterize biologically relevant changes in molecular signaling pathways among two groups, we furthermore calculated the enrichment for each pathway to identify significantly enriched concepts. All other parameters were set to default values. A nominal P<0.01 and FDR<0.25 were used as thresholds for determining the significance of the Enrichment Score (ES) [26,27].
Results
The expression landscape of PTPN2 in pancancer TCGA data
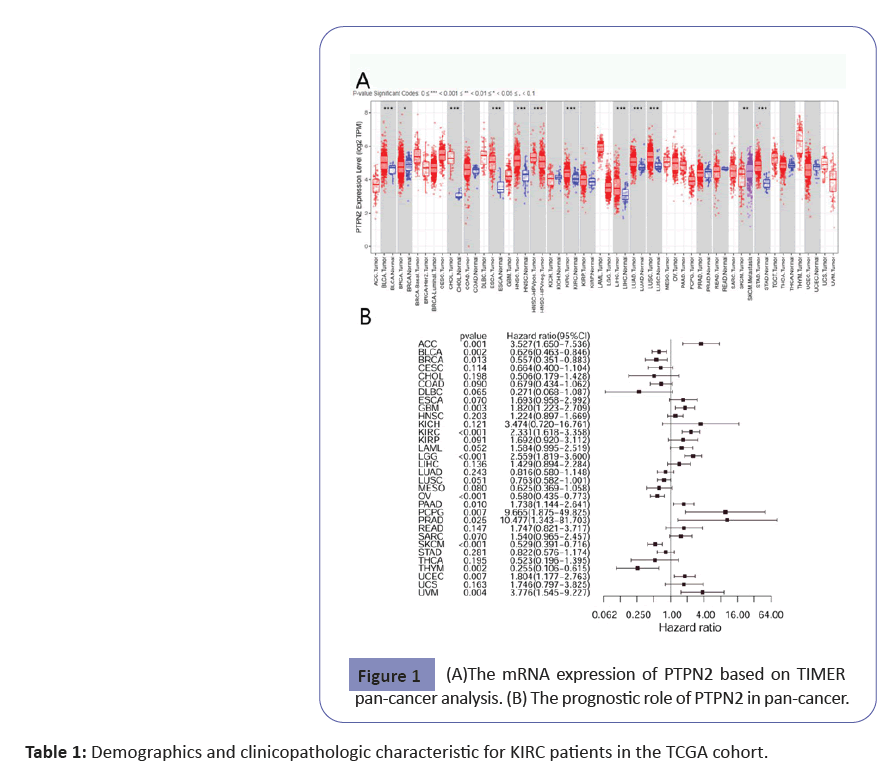
Integrated analysis was performed on tumor patients in The Cancer Genome Atlas (TCGA) datasets, a comprehensive database containing 11,000 patients’ samples. The expression profiling of PTPN2 was visualized by the TIMER platform based on pan-cancer TCGA data, including 31 different tumor types. Among 31 types of cancer, the result showed that PTPN2 was expressed in all kinds of cancer, but the expression levels were different. The highest expression was Thymoma (THYM), and the lowest was Liver Hepatocellular Carcinoma (LIHC). The expression level of other types of cancer is between these two types of cancer. Compared to normal tissue, PTPN2 was over-expressed in KIRC, LIHC, Lung Squamous Cell Carcinoma (LUSC), Stomach Adenocarcinoma (STAD), Head and Neck Squamous Cell Carcinoma (HNSC), Esophageal Carcinoma (ESCA), Bladder Urothelial Carcinoma (BLCA), and Cholangiocarcinoma (CHOL) based on TIMER pancancer analysis (Figure 1A).
we explored the relationship between mRNA expression of PTPN2 and patient survival using R. The relationship between PTPN2 and prognosis among different cancer types was summarized in Figure 1B. Among the 32 cancers examined, the high PTPN2 mRNA expression was found to be significantly correlated with decreased probability of survival for Adrenocortical Carcinoma (ACC) (HR=3.527; P=0.001), Glioblastoma Multiforme (GBM) (HR=1.820; P=0.003), KIRC (HR=2.331; P<0.001), Brain Lower Grade Glioma (LGG) (HR=2.559; P<0.001), Pancreatic Adenocarcinoma (PAAD) (HR=1.738; P=0.010), Pheochromocytoma and Paraganglioma (PCPG) (HR=9.665; P=0.007), Prostate adenocarcinoma (PRAD) (HR=10.477; P=0.025), Uterine Corpus Endometrial Carcinoma (UCEC) (HR=1.804; P= 0.007), and Uveal Melanoma (UVM) (HR=3.776; P=0.004). And high expression of PTPN2 correlated with better survival in patients with several types of cancer, including BLCA, Breast invasive carcinoma (BRCA), Ovarian serous cystadenocarcinoma (OV), Skin Cutaneous Melanoma (SKCM), and THYM.
The prognostic role of PTPN2 in KIRC
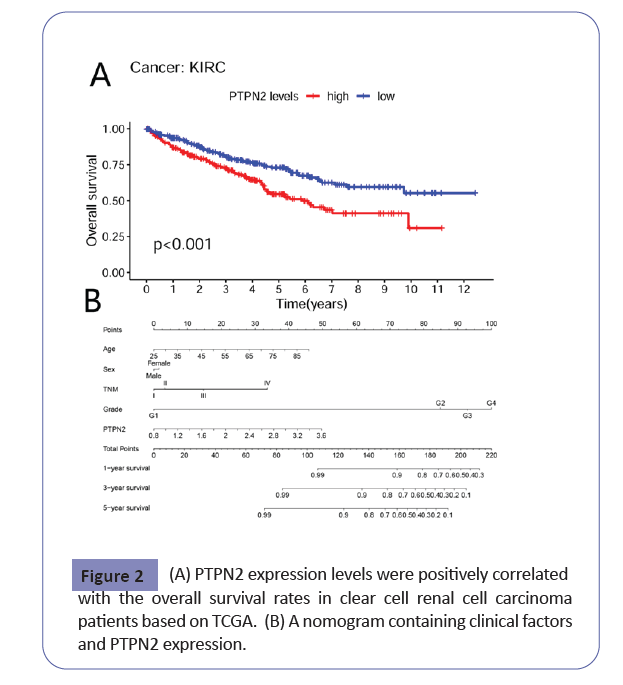
When we further investigate the potential clinical role of PTPN2 in KIRC patients, the results showed PTPN2 was closely related to the T stage (P=0.008), TNM stage (P=0.017), and Grade (P=0.002) as shown in Table 1. Overexpression of PTPN2 predicted poor survival in KIRC based on the TCGA cohort, which was revealed by the Kaplan-Meier method (P<0.001) (Figure 2A). In the multiple Cox analysis, PTPN2, age, and Grade were independent risk factors for OS. The univariate and multivariate analyses are listed in Table 2. Furthermore, we established a nomogram to predict the probability of OS in KIRC patients (Figure 2B). In this model, the Grade stage, TNM stage, age, and PTPN2 expressions have important effects on KIRC overall survival prediction.
| Total | PTPN2 low expression | PTPN2 high expression | P-value | |
|---|---|---|---|---|
| Age | 60.56 ± 12.14 | 60.59 ± 12.18 | 60.51 ± 12.10 | 0.939 |
| Sex | ||||
| Female | 186 | 99 | 87 | 0.372 |
| Male | 345 | 167 | 178 | |
| T stage | 0.008 | |||
| T1 | 272 | 155 | 117 | |
| T2 | 69 | 33 | 36 | |
| T3 | 200 | 98 | 102 | |
| T4 | 11 | 1 | 10 | |
| TNM | 0.017 | |||
| Stage I | 266 | 152 | 114 | |
| Stage II | 57 | 28 | 29 | |
| Stage III | 123 | 53 | 70 | |
| Stage IV | 82 | 32 | 50 | |
| NA | 3 | 1 | 2 | |
| M stage | 0.059 | |||
| M0 | 420 | 222 | 198 | |
| M1 | 78 | 31 | 47 | |
| Mx | 31 | 13 | 18 | |
| NA | 2 | 0 | 2 | |
| N stage | ||||
| N0 | 239 | 113 | 126 | 0.213 |
| N1 | 16 | 5 | 11 | |
| Nx | 276 | 148 | 128 | |
| Grade | 0.002 | |||
| G1 | 14 | 9 | 5 | |
| G2 | 227 | 116 | 111 | |
| G3 | 207 | 114 | 93 | |
| G4 | 75 | 23 | 52 | |
| Gx | 5 | 1 | 4 | |
| NA | 3 | 3 | 0 |
Table 1: Demographics and clinicopathologic characteristic for KIRC patients in the TCGA cohort.
| Characteristics | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Age | 1.029 | 1.016-1.042 | <0.001 | 1.027 | 1.009-1.046 | 0.003 |
| T stage | 1.892 | 1.606-2.230 | <0.001 | 0.872 | 0.540-1.407 | 0.574 |
| N stage | 3.15 | 1.626-6.101 | 0.001 | 1.427 | 0.698-2.196 | 0.33 |
| M stage | 4.256 | 3.108-5.830 | <0.001 | 1.521 | 0.676-3.419 | 0.311 |
| TNM stage | 1.857 | 1.625-2.123 | <0.001 | 1.584 | 0.944-2.659 | 0.081 |
| Grade | 2.244 | 1.828-2.754 | <0.001 | 1.48 | 1.067-2.053 | 0.019 |
| PTPN2 | 2.389 | 1.659-3.439 | <0.001 | 1.947 | 1.431-2.649 | <0.001 |
Table 2: Univariate and multivariate analyses of prognostic factors for overall survival using Cox proportional hazards regression model (N=531).
PTPN2 and TIICs
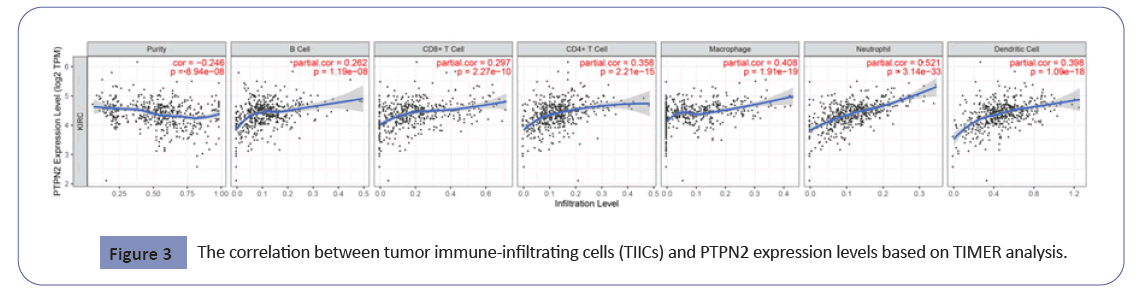
The Tumour Immunological Estimation Resource (TIMER) platform was explored the correlation between tumor Immune-Infiltrating Cells (TIICs) and PTPN2, and the statistical results were based on Spearman correlation analysis. The correlation between PTPN2 and TIICs was very prominent. PTPN2 was related to all six types of TIICs (B cells, P=1.19e-8; CD8+T cells, P=2.27e-10; CD4+T cells, P=2.21e-15; Macrophage, 1.19e-19; Neutrophils, P=3.14e-33; Dendritic cells, P=1.09e-18) (Figure 3).
Gene co-expression net analysis and GSEA analysis
To further investigate the underlying mechanism of PTPN2 in KIRC, gene co-expression net analyses were performed by Metascape. These co-expression genes were significantly enriched in mitotic cell cycle phase transition, centrosome duplication; DNA-dependent DNA replication; DNA repair; regulation of T cell activation . GSEA was performed here to identify the biological gene sets or pathways for PTPN2. we found 560 immune-related terms, including GSE29615_CTRL_ VS_DAY3_LAIV_IFLU_VACCINE_PBMC_UP; GSE1460_NAIVE_ CD4_TCELL_CORD_BLOOD_VS_THYMIC_STROMAL_CELL_DN; GSE12839_CTRL_VS_IL12_TREATED_PBMC_UP; SE21546_ UNSTIM_VS_ANTI_CD3_STIM_SAP1A_KO_DP_THYMOCYTES_ UP; GSE16385_UNTREATED_VS_12H_ROSIGLITAZONE_IL4_ TREATED_MACROPHAGE_DN
For gene set enrichment analysis, a series of GO terms related to immune and inflammatory factors were also found, including CD4 T-cell differentiation-related factors, innate and adaptive responses to vaccination, regulating lipid metabolism and inflammatory response in macrophages and dendritic cells and systemic inflammatory regulation related factors.
PTPN2 and immune-related genes
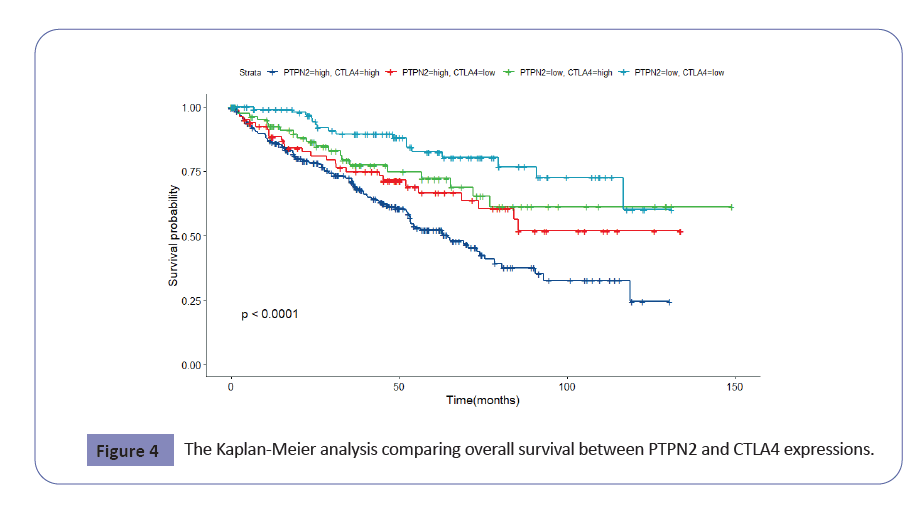
We acquired the immune-related genes from the ImmPort database. Then we identified the correlation between PTPN2 and these immune-related genes. The results showed that PTPN2 was related to many immune-related genes. To our greatest interest, CTLA-4 (P=8.31E-39, r=0.495) and PDCD1 (P=1.22E-14, r=0.306) were included in these related many immune-related genes. Furthermore, when the patients with KIRC were grouped into four strata by the level of PTPN2 and CTLA-4, the survival rate in patients with high PTPN2 and CTLA4 was significantly lower than those other three strata (Figure 4).
Discussion
We performed an integrative analysis focused on the expression of PTPN2 based on TCGA clinical tumor cases and identified the over-expression of PTPN2 in KIRC. Next, we found PTPN2 was a prognostic marker in KIRC. Furthermore, the multivariate analysis demonstrated that age, Grade, and PTPN2 were independent prognostic factors in TCGA KIRC dataset. More importantly, using Timer platform analysis, we found PTPN2 correlated with B cell, CD8+T cell, CD4+T cell, Macrophage, Neutrophil, and Dendritic cell. Then, CTLA4 was screened from IMMPORT data due to the results of correlation analysis. So, PTPN2 combined with CTLA4 as a model to predict the prognosis of KIRC was constructed; this model could help clinicians predict the disease prognosis and select the suitable treatment for KIRC.
PTPN2 was a protein tyrosine phosphatase family member, which was associated with many key signaling in tumorigenesis. PTPN2 was deleted in 6% of all T cell acute lymphoblastic leukemia, and it was involved in JAK/STAT signaling and tumorigenesis. In chronic myeloid leukemia patients, high PTPN2 expression was associated with poor Major Molecular Response (MMR). Shilds et al. reported that PTPN2 was deficient in triple-negative primary breast cancer [11]. They also found that loss of PTPN2 in the human breast cancer cell lines could increase cell proliferation. In our study, PTPN2 was over-expressed in KIRC, LIHC, LUSC, STAD, HNSC, ESCA, BLCA, and CHOL based on TIMER pan-cancer analysis. Targeting on PTPN2 may be a novel candidate gene for personalized tumour treatment in these cancer types. The study from Wang et al. also found PTPN2 expression level was increased in glioblastomas and associated with gliomas of the IDH wild-type and mesenchymal subtype. In 2017, PTPN2 was identified as a cancer immunotherapy target through CRISPR screening, which was published in Nature [11]. They also found that the increased sensitivity to anti-PD-1 immunotherapy in PTPN2-deficient tumors dependent on IFN-γ signalling [11]. These findings redefine our understanding of the role of PTPN2 in the tumor [13-18].
Immune checkpoint inhibitors have been successfully used in various solid tumors, such as lung cancer and breast cancer. The combination of immunosuppressive agents has a good effect on these types of tumors, such as anti-CTLA-4 combine anti-PD-1. In our study, we not only found PTPN2 was correlated with many immune cells but also screened out CTLA-4, which was associated with PTPN2. Similarly, to sensitize anti-PD-1 immunotherapy, our findings suggested that if PTPN2 deletion may also sensitize tumors to anti-CTLA-4 immunotherapy [19-22].
Conclusion
In conclusion, high PTPN2 levels significantly correlated with poor survival in ccRCC. PTPN2 is extremely closely associated with many types of TILs and CTLA-4. Our comprehensive bioinformatics analysis results, PTPN2 may be a potential prognosis biomarker and a novel immunotherapeutic target for KIRC.
Acknowledgements
The authors gratefully acknowledge the Cancer Genome Atlas (TCGA) database, which made the data available.
Funding
None.
Availability of Data and Materials
The raw data of this study are derived from the TCGA database, which are publicly available databases.
Ethics Approval and Consent to Participate
Not necessary.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Author’s Contributions
LP is the principle investigator. LP conducted statistical analysis and data management and edited and revised the manuscript.
References
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, et al. (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136: E359-386.
- Gupta K, Miller JD, Li JZ, Russell MW, Charbonneau C (2008) Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat Rev 34: 193-205.
- Scherr AJ, Lima JP, Sasse EC, Lima CS, Sasse AD (2011) Adjuvant therapy for locally advanced renal cell cancer: a systematic review with meta-analysis. BMC Cancer 11: 115.
- Hammers HJ, Plimack ER, Infante JR, Rini BI, Lewis LD, et al. (2017) Safety and Efficacy of Nivolumab in Combination With Ipilimumab in Metastatic Renal Cell Carcinoma: The CheckMate 016 Study. J Clin Oncol 35: 3851-3858.
- Rini BI, Atkins MB (2009) Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol 10: 992-1000.
- Braun DA, Bakouny Z, Hirsch L, Flippot R, Choueiri TK et al. (2021) Beyond conventional immune-checkpoint inhibition - novel immunotherapies for renal cell carcinoma. Nat Rev Clin Oncol 18: 199-214.
- Cool DE, Tonks NK, Charbonneau H, Walsh KA, Fischer EH, et al. (1989) cDNA isolated from a human T-cell library encodes a member of the protein-tyrosine-phosphatase family. Proc Natl Acad Sci U S A 86: 5257-5261.
- Pike KA, Tremblay ML (2016) TC-PTP and PTP1B: Regulating JAK-STAT signaling, controlling lymphoid malignancies. Cytokine 82: 52-57.
- Karlsson E, Veenstra C, Emin S, Dutta C, Perez-Tenorio G, et al. (2015) Loss of protein tyrosine phosphatase, non-receptor type 2 is associated with activation of AKT and tamoxifen resistance in breast cancer. Breast Cancer Res Treat 153: 31- 40.
- Grohmann M, Wiede F, Dodd GT, Gurzov EN, Butt T, et al. (2018) Obesity Drives STAT-1-Dependent NASH and STAT-3- Dependent HCC. Cell 175: 1289-1306.e1220.
- Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, et al. (2017) In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547: 413-418.
- Wiede F, Ziegler A, Zehn D, Tiganis T (2014) PTPN2 restrains CD8(+) T cell responses after antigen cross-presentation for the maintenance of peripheral tolerance in mice. J Autoimmun 53: 105-114.
- Morales LD, Archbold AK, Olivarez S, Slaga TJ, Kim DJ (2019). The role of T-cell protein tyrosine phosphatase in epithelial carcinogenesis. Mol Carcinog 58: 1640-1647.
- Kleppe M, Lahortiga I, El Chaar T, Mentens N, Graux G, et al. (2010) Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat Genet 42: 530-535.
- Kleppe M, Soulier J, Asnafi V, Mentens N, Knoops L, et al. (2011 ) PTPN2 negatively regulates oncogenic JAK1 in T-cell acute lymphoblastic leukemia. Blood 117: 7090-7098.
- Kok CH, Leclercq T, Watkins DB, Saunders V, Wang J, et al. (2014) Elevated PTPN2 expression is associated with inferior molecular response in de-novo chronic myeloid leukaemia patients. Leukemia 28: 702-705.
- Shields BJ, Wiede F, Gurzov EN, Hauser WC, Zhu HJ, et al. (2013) TCPTP regulates SFK and STAT3 signaling and is lost in triple-negative breast cancers. Mol Cell Biol 33: 557-570.
- Wang PF, Cai HQ, Zhang CB, Li YM, Liu X et al. (2018) Molecular and clinical characterization of PTPN2 expression from RNA- seq data of 996 brain gliomas. J Neuroinflammation 15: 145.
- Uruga H, Mino-Kenudson M (2021) Predictive biomarkers for response to immune checkpoint inhibitors in lung cancer: PD-L1 and beyond. Virchows Arch 478: 31-44.
- Setordzi P, Chang X, Liu Z, Wu Y, Zuo D (2021) The recent advances of PD-1 and PD-L1 checkpoint signaling inhibition for breast cancer immunotherapy. Eur J Pharmacol 895: 173867.
- Yang Y, Jin G, Pang Y, Hyuang Y, Wang W, et al. (2020) Comparative Efficacy and Safety of Nivolumab and Nivolumab Plus Ipilimumab in Advanced Cancer: A Systematic Review and Meta-Analysis. Front Pharmacol 11:40.
- Motzer RJ, Escudier B, McDermott DF, Frontera OA, Melichar B, et al. (2020) Survival outcomes and independent response assessment with nivolumab plus ipilimumab versus sunitinib in patients with advanced renal cell carcinoma: 42-month follow-up of a randomized phase 3 clinical trial. J Immunother Cancer 8: e000891.
- Li B, Severson E, Pignon JC, Zhao H, Li T, et al. (2016) Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome biology 17: 174.
- Zhou Y, Zhou B, Pache L, Chang M, Chanda Sk et al. (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10: 1523.
- Bhattacharya S, Andorf S, Gomes L, Schaefer H, Smith T, et al. (2014) ImmPort: disseminating data to the public for the future of immunology. Immunol Res 58: 234-239.
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, et al. (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267-273.
- Subramanian A, Tamayo P, Mootha VK, Benner C, Gillete MA et al. (2005) Gene set enrichment analysis: a knowledge- based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545-15550
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences