Identification of Differentially Expressed Genes in Urinary Bladder Cancer by Meta-Analysis by Using a Bioinformatics Tool
Mylsamy S, Devaki K
1Department of Biochemistry, CMS College of Science and Commerce, Coimbatore, Tamil Nadu, India
2Department of Biochemistry, Karpagam Academy of Higher Education, Coimbatore, Tamil Nadu, India
- Corresponding Author:
- Sathya Mylsamy
CMS College of Science and Commerce, Tamil Nadu, India
Tel: (+91)7904349498
E-mail: sathya268@redffmail.com
Received date: November 18, 2020 ; Accepted date: Decem ber 02, 2020 ; Published date: December 08, 2020
Citation: Mylsamy S, Devaki K (2020) Identification of Differentially Expressed Genes in Urinary Bladder Cancer by Meta-Analysis by using a Bioinformatics Tool. J Clin Exp Nephrol Vol.5 No.5:95.
Copyright: © 2020 Mylsamy S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: Bladder cancer is the ninth most prevalent malignant disease globally, which ranges from mild with low mortality rate to extremely high grade tumors associated with high mortality rate. The present study was aimed to identify the key genes associated with bladder cancer progression and later it may also be used as marker in the diagnosis and prognosis.
Materials and methods: The GSE3167, GSE7476, GSE68928 and GSE31189 data expression profiles were downloaded from the Gene Expression Omnibus (GEO) which includes 124 bladder cancer samples and 66 normal bladder tissues. The MGEx-TDB tool was used to analyse and find out the differentially expressed genes (DEGs), and the Gene Ontology (GO) functional annotation and KEGG pathway analysis were performed. Protein-protein Interaction (PPI) network ad based on centrality analysis hub genes was identified and were analysed towards in the diagnosis and prognosis of bladder cancer.
Results: In total, 475 differentially expressed genes including 196 up regulated genes and 279 down regulated genes were obtained from the four data sets analysis. GO analysis of the DEGs revealed that the up regulated genes were associated with mitotic nuclear division, cell division and apoptotic process. The down regulated genes were coupled with cell adhesion, MAPK cascade, cell differentiation. KEGG pathway analysis has shown that the up regulated genes were enriched in the pathways such as cell cycle, p53 signaling pathway and FoxO signaling pathway. The down regulated genes were enriched in pathways such as Focal adhesion, MAPK signaling pathway, platelet activation, proteoglycans in cancer, and pathways in cancer. From the constructed PPI network, based on higher degree, the hub genes were identified. CDK1, CCNB2, MAPK14, CDC20, STAT1 and NUSAP1 from the up regulated genes and FYN, UBC and ADAM22 from the down regulated genes.
Conclusion: This study enabled us to identify the key genes and the associated pathways. This will help us to understand the mechanism behind the tumor progression and its diagnosis.
Keywords
Bladder cancer; GEO; CCNB2; NUSAP1; ADAM22; KEGG; Gene ontology
Introduction
The bladder is probably one of the few sites in the body where the environmental factors play an undeniable role in genesis of cancer [1]. Carcinoma of the urinary bladder, the fourth most common cancer in men and the ninth most common cancer in women, with more than 400,000 new cases diagnosed every year especially in males and elderly and it is a disease ranges from mild with low mortality rate to extremely high grade tumors associated with high mortality tumors [2,3]. The survival rate of bladder carcinoma sharply declined with the spreading of the tumor and several studies have made known that various risk factors that may induce BC, including geography, race, gender, schistosomiasis infection, environmental or occupational exposure, smoking and genetic susceptibility [4].
Bladder cancer is hard to diagnose because the symptoms are unspecific: such as irritative voiding and painless hematuria and the gold standard for the diagnosis of bladder cancer is cystoscopy and urine cytology. Cystoscopy is invasive, complicated to perform and expensive, and makes patients feel uncomfortable; urine cytology involves the use of exfoliated cells in excreted urine to diagnose bladder cancer and has a low sensitivity for low-grade tumors [5,6].
Bladder cancer progression is a complicated and a multistep process involving changes both at molecular and genetic levels including changes in various genes, oncogenes, cell – cycle associated genes, tumor suppressor genes and DNA damage repair genes [7,4]. Unravelling the biological complexity behind the bladder cancer will be advantageous towards providing novel tools for the early diagnosis, prognosis evaluation and recurrence monitoring, and the development of methods for controlling the proliferation of bladder cancer cells and identification of therapeutic targets by selecting those molecular targets significantly expressed in bladder tumors [8]. Global profiling of gene expression by microarray technology is widely used to study molecular mechanism of cancer [9].
controlling the proliferation of bladder cancer cells and identification of therapeutic targets by selecting those molecular targets significantly expressed in bladder tumors [8]. Global profiling of gene expression by microarray technology is widely used to study molecular mechanism of cancer [9].
Meta-analysis which merges all qualified datasets into a single analysis using a more robust statistical method is preferable to yield a more meaningful set of differentially expressed genes and to provide new sights into the biological mechanisms [4].
The identification of differentially regulated genes and gene networks allows the dissection of pathways and processes that are dysregulated in bladder cancer. This, in turn, provides invaluable clues relating to pathogenesis and moreover, may provide novel targets for drug development [11].
In the present study, we conducted a meta-analysis of 4 datasets retrieved from GEO to investigate the molecular and genetic mechanisms behind the bladder cancer.
Materials and Methods
Collection of microarray data and its processing
We systematically retrieved the data from Gene Expression Omnibus (GEO): A microarray data repository, which was queried with bladder and different cancer related terms (e.g., cancer, carcinoma, and sarcoma). The hits were then manually screened to obtain relevant studies. The following criteria were used for determining the relevance. The datasets were selected based on the inclusion and exclusion criteria
Inclusion Criteria for Samples/Studies: Study should have profiled human bladder cancer and normal bladder tissue samples. Sample considered in the study should be a primary tumor. Study should have been performed using Affymetrix microarray platforms.
Exclusion Criteria for Samples/Studies: Studies related to cell line or cultured cells Studies related to treatments such as, drug and chemical treatment; transfection etc. studies related to xenografts were excluded.
Based on the above mentioned criteria, four datasets with the accession number GSE3167, GSE7476, GSE31189 and GSE68928 were selected for the present study. These datasets were having total of 190 samples (124 bladder cancer and 66 normal bladder samples).
Identification of Differentially Expressed Genes (DEGs)
In the present study, the DEGs between bladder cancer samples and normal bladder samples were analysed by using a method published by Acharya et al. The approach has been successfully used to obtain candidate genes in various normal and disease conditions of testis and uterus tissues, and development of gene expression databases for testis. The DEGs that were expressed in bladder carcinoma relative to the control groups were selected based on the cut off criteria value of log fold change FC>2 and P<0.05.
Gene Ontology (GO) analysis
Gene Ontology analysis is a common useful approach for annotating genes and gene products and for identifying characteristic biological phenomena for high throughput genome or transcriptome data. To describe gene product attributes, GO provides three categories of defined terms, including biological process (BP), cellular component (CC) and molecular function (MF) categories. Significant genes were considered for functional analysis, using DAVID functional annotation tool. Pathways and GO annotations having a p value of at least 0.05 were considered significant [12].
Pathway enrichment analysis
To investigate the enriched signaling pathways of DEGs, pathway enrichment analysis was done using KEGG database (www.genome.jp/KEGG). This analysis is useful for investigating if the differentially expressed genes are associated with certain set of genes and pathways. All of the metabolic and nonmetabolic pathways available from KEGG database were used as DAVID inputs for analysis. A value of P<0.1 and at least seven DEGs were chosen as cut off criteria [13].
Protein-Protein Interaction (PPI) network
The information about protein-protein interaction is highly useful for the users and it should be easily available in web and should be easily accessible for the users. The Search Tool for the Retrieval of Interacting Genes (STRING) database resources aims to provide this service by acting as a “one-stop shop” for all information on functional links between proteins. STRING is the only site which covers the interaction map of hundreds more than 1100 organisms ranging from single cell organisms to humans.
Centrality analysis for identifying the Hub genes
Centrality analysis was performed to identify the key genes in the bladder carcinoma based on the nodes degree in the protein interaction network [14]. Centrality analysis mainly contain degree centrality, closeness centrality and shortest path between centrality, in which degree the equivalent of the number of nodes directly adjacent to a given node (indicating the degree vertex), is a simplest topological index.
In the present study, based on centrality analysis the hub gens involved in biological process were identified from the PPI networks. Nodes with high degree (highly connected) are called Hubs which interconnect with several other genes signifying a central role in the interaction network. Genes with degree>15 were defined as Hub gene for the present study [14,2].
Results and Discussion
Identification of Differentially Expressed Genes (DEGs)
Based on the inclusion and exclusion criteria, meta-analysis of the four selected microarray data was done using an on-line tool MGEx-Tdb. The selected four studies contained a total of 190 samples which includes 124 bladder cancer and 66 normal bladder samples. Based on the analysis, a total of 475 differentially expressed genes were identified in the urinary bladder cancer samples compared with normal urothelium. Based on the genes having average p value of less than 0.05, minimum average fold change 1.2 (log 2 fold of 0.26) and minimum differential reliability score 4, out of 475 dysregualted genes, 196 genes were found as up regulated and 279 genes as down regulated in comparison with normal urothelium.
Gene Ontology (GO) analysis of differentially expressed genes
Functional classification of DEGs at a statistical cut off criterion of p<0.001 using an online tool DAVID (Database for Annotation, visualization and integrated Discovery) indicated significant enrichment of these genes in various categories of GO (Biological processes, cellular components and Molecular function). Out of the 196 up regulated genes, 27 were involved in biological process, 12 in molecular function, and 19 in cellular components.
The up-regulated genes were significantly involved in biological processes associated with mitotic nuclear division, cell division, sister chromatid cohesion, chromosome segregation, cell proliferation, apoptotic process, positive regulation of apoptotic process, while the down regulated genes were mainly enriched in cell adhesion, MAPK cascade, negative regulation of cell migration, negative regulation of transcription from RNA polymerase II promoter.
GO cellular component (CC) analysis showed that the up regulated DEGs were significantly enriched in cytosol, nucleoplasm, cytoplasm, membrane, nucleus and nucleolus and the down regulated DEGs were enriched in synapse, cytoplasm and cytosol. In addition, the molecular function of up-regulated DEGs were mainly associated with protein binding and ATP binding, while down regulated DEGs were involved in calmodulin binding and actin binding (Table 1).
| Category | Term | Count | P value |
|---|---|---|---|
| Up regulated | |||
| BP | |||
| GO:0007067 | Mitotic nuclear division | 19 | 3.15E-11 |
| GO:0051301 | Cell division | 17 | 2.86E-07 |
| GO:0007062 | Sister chromatid cohesion | 10 | 6.70E-07 |
| GO:0007059 | Chromosome segregation | 8 | 4.04E-06 |
| GO:0008283 | Cell proliferation | 11 | 0.002961 |
| GO:0006915 | Apoptotic process | 12 | 0.021711 |
| GO:0043065 | Positive regulation of apoptotic process | 10 | 0.0029613 |
| Down regulated | |||
| GO:0007155 | Cell adhesion | 16 | 0.001567494 |
| GO:0000165 | MAPK cascade | 10 | 0.00973433 |
| GO:0030336 | Negative regulation of cell migration | 8 | 3.11E-04 |
| GO:0030154 | Cell differentiation | 13 | 0.024333194 |
| CC | |||
| Up Regulated | |||
| GO:0005829 | Cytosol | 62 | 3.02E-08 |
| GO:0005654 | Nucleoplasm | 53 | 3.26E-07 |
| GO:0005737 | Cytoplasm | 70 | 5.30E-04 |
| GO:0016020 | Membrane | 36 | 0.001029 |
| GO:0005634 | Nucleus | 66 | 0.010298 |
| Down regulated | |||
| GO:0045202 | Synapse | 19 | 5.48E-04 |
| GO:0043025 | Neuronal cell body | 16 | 0.00752722 |
| MF | |||
| Up regulated | |||
| GO:0005515 | Protein Binding | 110 | 2.44E-05 |
| GO:0005524 | ATP binding | 26 | 0.04408 |
| Down regulated | |||
| GO:0005516 | Calmodulin binding | 20 | 8.46E-04 |
| GO:0003779 | Actin binding | 17 | 0.003596742 |
Table 1: Gene Ontology analysis of Differentially expressed genes.
Pathway enrichment analysis
To gain more insights of the changes at a functional level, pathway analysis were done for up regulated genes according to KEGG analysis using an online tool DAVID with a P<0.05. The up regulated genes were essentially abundant in cancer related pathways such as cell cycle (hsa0411), including the genes (CCNB2, CDK1, PLK1, TP53, MAD2L1,MCM2, CDK2, CDC20, BUB1B, ATR etc.), p53 signaling pathway (hsa04115) with the genes (CDK4, CDK2,CCNB2, CDK1, CCNB1, TP53 etc.,), FoxO signaling pathway(hsa04068) with the genes(CDK2, CCNB2, MAPK, MDM2, STAT3, PTEN, PI3K, JNK, CK1, BCL-6). Table 2 gives the most significantly enriched KEGG pathways of up regulated genes and down regulated genes.
| Pathway ID | Term | Count | P-value | Genes |
|---|---|---|---|---|
| Up regulated | ||||
| hsa04110 | Cell cycle | 7 | 0.001653 | CCNB2, CDK1, PLK1, TP53MAD2L1, MCM2, CDK2, CDC20, ATR, PCNA, RAD21 BUB1B, CDC6CCNE2, PCNA, CCNA2, CDC6 |
| hsa04115 | p53 signaling pathway | 5 | 0.004819 | CDK4, CDK2, CCNE2, CHEK2, CCNB2, CDK1, CCNB1, CASP3, TP53, RM2, GADD45A, PMA1P1 |
| hsa04068 | FoxO signaling pathway | 5 | 0.048488 | CDK2, CCNB2, MAPK, MDM2, STAT3, PTEN, PI3K, JNK, CK1, BCL-6,SGK1, SKP2, GADD45A |
| Down regulated | ||||
| hsa04510 | Focal adhesion | 13 | 1.27E-05 | ITGA3, VCL, MAPK1, PXN, MYL2, VASP, RAP1B, RAF1THBS1, ITGAV, CCND2 |
| hsa05414 | Dilated cardiomyopathy | 6 | 0.004611499 | ITGA3, MYL2, ITGAV, ITGB7, ITGB5, PRKACA, ADCY2, PLN, ITGA 8, ATP2A2 |
| hsa04010 | MAPK signaling pathway | 10 | 0.005724748 | MAPK1, RAP1B, RAF1, GADD45G, PDGFRB, AKT3, RAPGEF2, TP53, FGF18, PAK1 |
| hsa04810 | Regulation of actin cytoskeleton | 9 | 0.005993404 | ITGA3, VCL, MAPK1, WASL, PXN, MYL2, RAF1, ITGAV, PDGFRB, PIK3CA, ITGB7, PAK1 |
| hsa04611 | Platelet activation | 7 | 0.00677811 | RHO, TP, G13, PKA, MLCK, PI3K, GPV |
| hsa04520 | Adherens junction | 5 | 0.01347986 | VCL, MAPK1, WASL, CDH1, CDC42, SMAD3, SSX2IP, CTNNB1, FYN, SRC, SORBS1 |
| hsa05205 | Proteoglycans in cancer | 8 | 0.015094979 | FGFR1, IGF1, SDC2, JIMP3, DCN, ANK2, RRAS, FAS, FGF2, CAV1, CAV2, LUM, FZO7 |
| hsa04270 | Vascular smooth muscle contraction | 6 | 0.019157727 | MAPK1, ACTA2, RAF1, ITPR1, PRKACA, ADCY2, CALM1, MYLK, CALD1, MYH11, ROCK1, RHO |
| hsa05200 | Pathways in cancer | 11 | 0.030976077 | DVL2, ITGA3, MAPK1, PLCG1, RAF1, VHL, ITGAV, PDGFRB, CREBBP, EP300, AKT3, PIK3CA |
| hsa04530 | Tight junction | 6 | 0.032714483 | MYL2, AKT3, HRAS, CDC42, CTNNB1, MAGI2 |
| hsa04310 | Wnt signaling pathway | 6 | 0.03366636 | DVL2, CCND2, AXIN1, CREBBP, EP300, TP53, CTBP1, PRICKLE2, AXIN2, PRKACA, CTBP2, PSEN1 |
Table 2: KEGG Pathway Analysis of DEGs.
Protein-Protein Interaction (PPI) network construction and Hub gene/protein screening
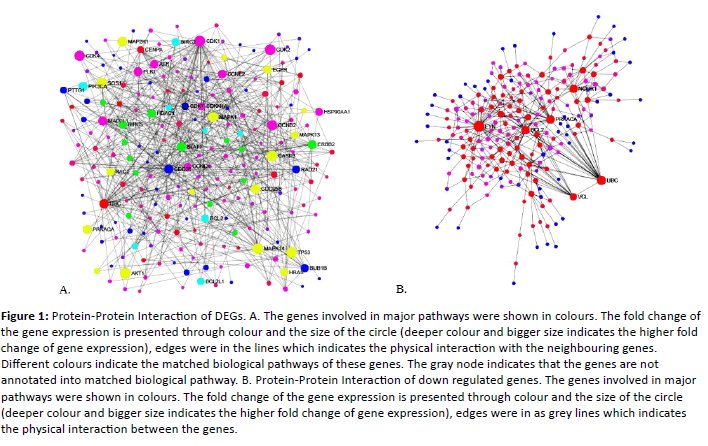
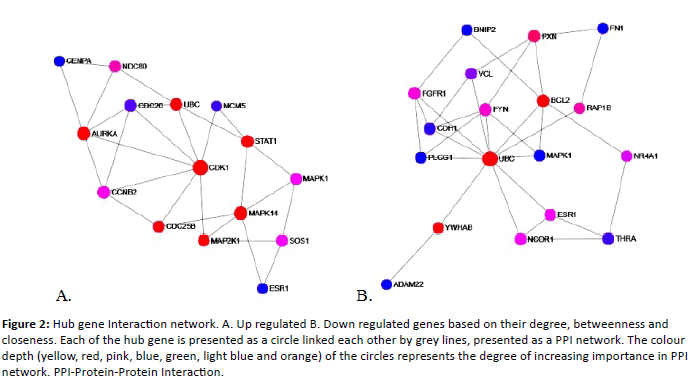
In bladder carcinoma 475 genes were dysregualted when compared to the normal urothelium. Out of 475, 196 genes were up regulated and 275 were down regulated. To systematically analyse the various functions of DEGs in bladder carcinoma, these DEGs mapped to Protein-Protein Interaction (PPI) network using a database resource Search Tool for the Retrival of Interacting Genes (STRING). Interacting pairs with a combined score more than>0.9 were considered to be significant and employed to construct the networks. Based on centrality analysis, the genes having higher degree are termed as hub genes/proteins. In the present study the genes having>20 degrees were screened out from the PPI network and those were given in the Table 3. Figure 1 represents the PPI network of up regulated and down regulated DEGs. The PPI interaction among the hub genes were given in Figure 2.
Figure 1: Protein-Protein Interaction of DEGs. A. The genes involved in major pathways were shown in colours. The fold change of the gene expression is presented through colour and the size of the circle (deeper colour and bigger size indicates the higher fold change of gene expression), edges were in the lines which indicates the physical interaction with the neighbouring genes. Different colours indicate the matched biological pathways of these genes. The gray node indicates that the genes are not annotated into matched biological pathway. B. Protein-Protein Interaction of down regulated genes. The genes involved in major pathways were shown in colours. The fold change of the gene expression is presented through colour and the size of the circle (deeper colour and bigger size indicates the higher fold change of gene expression), edges were in as grey lines which indicates the physical interaction between the genes.
Figure 2: Hub gene Interaction network. A. Up regulated B. Down regulated genes based on their degree, betweenness and closeness. Each of the hub gene is presented as a circle linked each other by grey lines, presented as a PPI network. The colour depth (yellow, red, pink, blue, green, light blue and orange) of the circles represents the degree of increasing importance in PPI network. PPI-Protein-Protein Interaction.
| Id | Label | Degree | Betweenness | Closeness |
|---|---|---|---|---|
| Up regulated | ||||
| ENSP00000378699 | CDK1 | 49 | 7469.24 | 0.00025323 |
| ENSP00000344818 | CCNB2 | 44 | 10067.4 | 0.00022883 |
| ENSP00000229794 | MAPK14 | 25 | 2913.01 | 0.0002061 |
| ENSP00000308450 | CDC20 | 21 | 737.29 | 0.00024814 |
| ENSP00000354394 | STAT1 | 20 | 3584.04 | 0.0002343 |
| ENSP00000269571 | NUSAP1 | 20 | 2572.68 | 0.00018563 |
| Down regulated | ||||
| ENSP00000346671 | FYN | 39 | 8426.15 | 0.00031075 |
| ENSP00000344818 | UBC | 25 | 9427.52 | 0.00027655 |
| ENSP00000309591 | ADAM22 | 24 | 3230.09 | 0.00027663 |
Table 3: Hub genes/proteins.
Bladder cancer displays a great deal of heterogeneity in comparison to many other types of cancer and also exhibits a wide spectrum of clinical and pathological features [3,14].
About 70-80% of bladder cancer is diagnosed as non-muscle invasive bladder cancer (NMIBC) and 20-30% as muscle invasive bladder cancer (MIBC). 10-30% of patients with NMIBC may progress to MIBC. Early detection and early diagnosis is most needed in the bladder cancer treatment. Urine cytology and cytoscopy are the gold standard in the diagnosis of bladder cancer. They are often accompanied by several adverse effects. Thus, there is an urgent need to develop novel diagnostic methods for the early detection and treatment [6]. Gene expression profiling by microarray powerful approach to identify potential biomarkers involved in the disease progression and widely adopted in diagnosis and therapeutic targets for bladder cancer [12].
In the present study, the data’s were extracted from GSE3167, GSE7476, GSE31189 and GSE68928 and 196 up regulated and 279 down regulated DEGs between bladder cancer and control specimens were identified using MGx-Tdb tool. The up regulated genes were enriched in cell cycle, p53 signaling pathway and FoxO signaling pathway, while the down regulated genes were mainly involved in focal adhesion, MAPK signaling pathway, regulation of actin cytoskeleton, platelet activation, adherens junction, proteoglycans in cancer, pathways in cancer and tight junctions. The PPI construction using DEGs given away the key genes in the bladder cancer formation and progression, which may be useful in future to understand the bladder cancer. Notably the key genes include CCNB2, CDK1, NUSAP1 and ADAM22 which may have a specific contributions to bladder cancer development to the progression and diagnosis.
CCNB2 gene (Cyclin B2), member of cyclin family proteins, which regulates the activities of cyclin dependant kinases (CDKs) and different cyclins function spatially and temporarily in specific phases of the cell cycle and numerous studies have shown that CCNB1 is over expressed and promotes tumor proliferation in a variety of tumors, such as breast cancer, colorectal cancer and hepatocellular carcinomas [15]. Vasserur et al., accounts that the two cyclins were associated with mammalian cells and both combine with CDK1, and its level will go up during G2 and peak in mitosis (M phase). Lei et al. reported that the up regulation of Cyclin B2 in urinary bladder cancer plays an important role and was correlated with poor cancer differentiation. The up regulation probably responsible for cell growth, invasion and migration in bladder cancer. The decrease in cyclin B2 expression in bladder cancer has little effect on cell growth but significantly inhibited cell invasion and migration and prolonged the survival times of nude mice in vivo and Cyclin B2 protein level was higher in bladder cancer tissues than that in normal tissues and higher in invasive cancers than that in non-invasive cancer [16]. Yuan et al., reported that the stronger expression of cyclin B2 mRNA in tumor cells was an independent predictor of a poor prognosis in patients with Adenocarcinoma of the lung, and CCNB2 may also function as an oncogene and could serve as a potential biomarker in breast cancer [17].
NUSAP 1 (Nucleoar Spindle Associated Protein 1) is a recently identified protein that plays a vital role in accurate chromosome segregation fidelity and is a therapeutic target in cancer and is over expressed in numerous cancers and high levels were correlated with poor prognosis in aggressive breast cancer. Li et al., 2017 reported as NUSAP1 regulates mitosis and high expression of NUSAP1 is involved in the progression of prostate cancer [14].
Fang et al., 2016 reported that NUSAP1 exhibits a cell cycle dependent localization and is selectively expressed in proliferating cells. Its mRNA and protein expression levels reach a peak at the transitions of G2/M and then rapidly decline after cell division. The depletion and over expression of NUSAP1 in cells result in abnormal chromosome segregation, aberrant spindle assembly, defective cytokinesis, G2/M arrest and microtubule bundling respectively.
ADAM22 (A Disintegrin and Metalloproteinase) a cell surface proteins, having highly conserved cystenine residues. STRING and Gene mania interactions reports shows that it is in interaction with LGI1 (Leucine–Rich Glioma Inactivated 1 ligand). Gene ontology analysis of down regulated genes reports that the changes are mostly with biological process such as modulations of synaptic transmission, axon guidance, synaptic vesicle clustering, epithelial cell-cell adhesion, cell differentiation, cell adhesion, etc, and the cellular process with neuronal cells. Fukata et al., reports that LGI1 is a neuronal peptide that has been shown to be a specific extracellular ligand for ADAM22 in the CNS [18,19]. Sagane et al., says LGI1 binds the extracellular domain of ADAM22 and may be functioning by inhibiting the disintergin domain. Doherty et al., reported that ADAM22 expression promotes both migration and dedifferentiation, key hall marks of metastasis. While clinically elevated ADAM22 expression has poor diseases free survival [20-23].
Conclusion
The dysregualted expression of CCNB2, NUSAP1 and ADAM22 play a vital role in G2/M transitions and promotes the mitotic cell division. These genes have little effect on growth it can not only be used as marker but also may be used as a therapeutic target either individually or in combination.
References
- P. Chinnaswamy (2004) Tumor markers recommended for diagnosis of carcinoma of the urinary bladder in health screening programmes. Wide spectrum 1: 10-14.
- Shariat SF, Karam JA, Lotan Y, Karakiewizc PI (2008) Critical evaluation of urinary markers for bladder cancer detection and monitoring. Rev Urol 10: 120.
- Giulietti M, Occhipinti G, Righetti A, Bracci M, Conti A, et al. (2018) Emerging biomarkers in bladder Cancer identified by network analysis of transcriptomic data. Front Oncol 8: 450.
- Zhang DQ, Zhou CK, Chen SZ, Yang Y, Shi BK (2017) Identification of hub genes and pathways associated with bladder cancer based on co-expression network analysis. Oncol Lett 14: 1115-1122.
- Vinayata Manuballa (2005) Biomarker p53: Role Towards Prognosis of Bladder Cancer. MMG 445 Basic Biotechnology eJournal.
- Gao X, Chen Y, Chen M, Wang S, Wen X, et al. (2018) Identification of key candidate genes and biological pathways in bladder cancer. Peer J 6: e6036.
- Liu M, Qiu YL, Jin T, Zhou Y, Mao ZY, et al. (2018) Meta-analysis of microarray datasets identify several chromosome segregation-related cancer/testis genes potentially contributing to anaplastic thyroid carcinoma. Peer J 6: e5822.
- Sanchez-Carbayo M, Socci ND, Lozano JJ, Li W, Charytonowicz E, et al. (2003) Gene discovery in bladder cancer progression using cDNA microarrays. Am J Pathol 163: 505-516.
- Qian X, Song X, He Y, Yang Z, Sun T, et al. (2015) CCNB2 overexpression is a poor prognostic biomarker in Chinese NSCLC patients. Biomed Pharmacother 74: 222-227.
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, et al. (2012) NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res 41: D991-995.
- Wu T, Zhang X, Huang X, Yang Y, Hua X (2010) Regulation of cyclin B2 expression and cell cycle G2/m transition by menin. J Biol Chem 285: 18291-18300.
- Yan M, Jing X, Liu Y, Cui X (2018) Screening and identification of key biomarkers in bladder carcinoma: Evidence from bioinformatics analysis. Oncol Lett 16: 30923-3100.
- Bolger JC, Young LS (2013) ADAM22 as a prognostic and therapeutic drug target in the treatment of endocrine-resistant breast cancer. Vitam Horm 93: 307-321.
- Li L, Lei Q, Zhang S, Kong L, Qin B (2017) Screening and identification of key biomarkers in hepatocellular carcinoma: evidence from bioinformatic analysis. Oncol Rep 38: 2607-2618.
- Mo ML, Chen Z, Li J, Li HL, Sheng Q, et al. (2010) Use of serum circulating CCNB2 in cancer surveillance. 236-242.
- Lei CY, Wang W, Zhu YT, Fang WY, Tan WL (2016) The decrease of cyclin B2 expression inhibits invasion and metastasis of bladder cancer. Urol Oncol Semin Ori 34: 237-e1.
- Yuan L, Chen L, Qian K, Qian G, Wu CL, et al. (2017) Co-expression network analysis identified six hub genes in association with progression and prognosis in human clear cell renal cell carcinoma (ccRCC). Genom data 14: 132-140.
- Fang Y, Yu H, Liang X, Xu J, Cai X (2014) Chk1-induced CCNB1 overexpression promotes cell proliferation and tumor growth in human colorectal cancer. Cancer boil ther 15: 1268-1279.
- Tang F, He Z, Lei H, Chen Y, Lu Z, et al. (2018) Identification of differentially expressed genes and biological pathways in bladder cancer. Mol Med Rep 17: 6425-6434.
- Nam HJ, Van Deursen JM (2014) Cyclin B2 and p53 control proper timing of centrosome separation. Nat cell biol 16: 535.
- Lei CY, Wang W, Zhu YT, Fang WY, Tan WL (2016) The decrease of cyclin B2 expression inhibits invasion and metastasis of bladder cancer. Urol Oncol: Seminars and Original Investigations 34: 237.
- Zhou L, Du Y, Kong L, Zhang X, Chen Q (2018) Identification of molecular target genes and key pathways in hepatocellular carcinoma by bioinformatics analysis. OncoTargets and ther 11: 1861.
- Li WM, Wei YC, Huang CN, Ke HL, Li CC, et al. (2016) Matrix metalloproteinase‐11 as a marker of metastasis and predictor of poor survival in urothelial carcinomas. J Surg Oncol 113: 700-707.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences